Recommended Publications for Intervention Outcome Prediction Models: Difference between revisions
Lorikrammer (talk | contribs) |
Lorikrammer (talk | contribs) |
||
| Line 7: | Line 7: | ||
All the genomic data was found at this link: https://www.cbioportal.org/study/summary?id=brca_tcga | All the genomic data was found at this link: https://www.cbioportal.org/study/summary?id=brca_tcga | ||
[[File:36358741 example table.png|left|frameless| | [[File:36358741 example table.png|left|frameless|601x601px]] | ||
| Line 15: | Line 15: | ||
<u>Curator comments:</u> I traced the link and looked at the summary page, which provided me with data on the cancer type, data types, and mutations. I focused on where I could find the response/nonresponse status. Since the response indicator for this PMID was “Patients had no disease progression after first-line chemotherapy during the 150 months follow-up period”, I looked for where I could find data on progression of the disease. I found this data on the KM plot for overall survival and disease free months. If we isolated each of the data points on this graph, we could get each patient ID, cancer ID, and other data that corresponds with the responder status of the ID. For example, I found that the data point for the patient ID TCGA-B6-A0I5 had the highest overall percentage disease free after 281 months. When I traced this ID on the clinical data tab, I found the specific mutations and genome alterations to the corresponding ID, which were two important factors looked at in the paper. | <u>Curator comments:</u> I traced the link and looked at the summary page, which provided me with data on the cancer type, data types, and mutations. I focused on where I could find the response/nonresponse status. Since the response indicator for this PMID was “Patients had no disease progression after first-line chemotherapy during the 150 months follow-up period”, I looked for where I could find data on progression of the disease. I found this data on the KM plot for overall survival and disease free months. If we isolated each of the data points on this graph, we could get each patient ID, cancer ID, and other data that corresponds with the responder status of the ID. For example, I found that the data point for the patient ID TCGA-B6-A0I5 had the highest overall percentage disease free after 281 months. When I traced this ID on the clinical data tab, I found the specific mutations and genome alterations to the corresponding ID, which were two important factors looked at in the paper. | ||
| Line 63: | Line 61: | ||
Detailed clinico-pathological information was collected from 6 different cohorts derived from public datasets excluding cases with additional chemotherapy or treatment. The different properties and metadata included in the datasets provided are in similar formats. With some filtering, the general dataset is presented as such: | Detailed clinico-pathological information was collected from 6 different cohorts derived from public datasets excluding cases with additional chemotherapy or treatment. The different properties and metadata included in the datasets provided are in similar formats. With some filtering, the general dataset is presented as such: | ||
[[File:27613525 example table.png|left|frameless|626x626px]] | [[File:27613525 example table.png|left|frameless|626x626px]] | ||
Then the two-gene classifier was calculated for the samples and sorted into low, medium, or high classifiers. 253 genes were selected based on literature research and array data was analyzed on patients with early stage lung cancer. | Then the two-gene classifier was calculated for the samples and sorted into low, medium, or high classifiers. 253 genes were selected based on literature research and array data was analyzed on patients with early stage lung cancer. | ||
Revision as of 20:16, 9 January 2026
The following publications have been evaluated for their usefulness in providing publicly-available datasets for intervention outcome prediction models.
Breast Cancer
PMID 36358741
The data was collected from the breast cancer cohort of The Cancer Genome Atlas (TCGA) database. The cohort size was 399 patients.
All the genomic data was found at this link: https://www.cbioportal.org/study/summary?id=brca_tcga
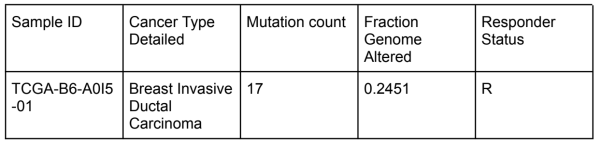
Curator comments: I traced the link and looked at the summary page, which provided me with data on the cancer type, data types, and mutations. I focused on where I could find the response/nonresponse status. Since the response indicator for this PMID was “Patients had no disease progression after first-line chemotherapy during the 150 months follow-up period”, I looked for where I could find data on progression of the disease. I found this data on the KM plot for overall survival and disease free months. If we isolated each of the data points on this graph, we could get each patient ID, cancer ID, and other data that corresponds with the responder status of the ID. For example, I found that the data point for the patient ID TCGA-B6-A0I5 had the highest overall percentage disease free after 281 months. When I traced this ID on the clinical data tab, I found the specific mutations and genome alterations to the corresponding ID, which were two important factors looked at in the paper.
PMID 36257316
The purpose of this study was to explore how Ferroptosis (a type of cell death linked to iron and lipid metabolism) works differently across triple-negative breast cancer (TNBC) tumors, and to test whether blocking the enzyme GPX4 could make TNBC cells more sensitive to immunotherapy (anti–PD-1).
Condition
- 465 TNBC patients from a large multi-omics dataset, including:
- 360 with transcriptomic data
- 279 with whole-exome sequencing (WES)
- 401 with somatic copy-number alteration (SCNA) data
- 330 with metabolomic data
- Additional validation done in LAR-like TNBC mouse models (TS/A cells in BALB/c mice).
- Clinical data came from the I-SPY2 cohort and related GEO datasets (GSE173839, GSE124821, GSE176078).
Intervention (drug treatment)
- Combination of GPX4 inhibitors (RSL3, ML162) and anti–PD-1 immunotherapy, tested both alone and together.
- Mice received treatments of:
- GPX4 inhibitor only
- anti–PD-1 only
- or the combination of both.
# of Patients / Samples
- 465 patients in the TNBC multi-omics cohort.
- 8 mice per treatment group in the in-vivo study.
Primary Endpoint
- To see whether GPX4 inhibition could:
- Slow down tumor growth.
- Change the tumor microenvironment to be more immune-active.
- Improve tumor control when combined with anti–PD-1 therapy.
Responder / Non-Responder Definition
Responders: Tumors where GPX4 inhibition caused ferroptosis, reduced growth, and made the tumor environment more inflammatory.
Non-Responders: Tumors with high GSH metabolism (glutathione pathway) that resisted ferroptosis and showed poor response to immunotherapy.
Lung Cancer
PMID 27613525
Detailed clinico-pathological information was collected from 6 different cohorts derived from public datasets excluding cases with additional chemotherapy or treatment. The different properties and metadata included in the datasets provided are in similar formats. With some filtering, the general dataset is presented as such:
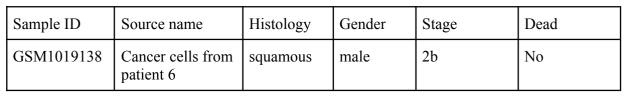
Then the two-gene classifier was calculated for the samples and sorted into low, medium, or high classifiers. 253 genes were selected based on literature research and array data was analyzed on patients with early stage lung cancer.
The results were validated using qRT-PCR in the same sample population and measured as significantly correlated (p < 0.05) with the microarray data for all of the 20 genes that were most significantly associated with relapse-free survival (RFS) using univariable Cox regression. 7 out of the 20 genes measured were significantly associated with RFS. Two of which analyzed were DUSP6 or ACTN4 in high expression indicated a worse prognosis in Japan and NCI-MD/Norway cohorts. This was validated with the other four cohorts and a fixed effects meta-analysis of the datasets demonstrated no heterogeneity or inconsistency. Therefore, the two-gene classifier reliably and precisely identified stage I + II and stage I patients at high risk for death.
Curator comments: The focus of the article is the predictive capability and potential of the two genes: DUSP6 and ACTN4 which can give an accurate result for the patient’s prognosis. Therefore, originally for the purposes of our study and the results of interventions, diagnosis biomarkers are not ideal, however, after looking at the datasets themselves, it would be interesting to consider its capability for our purposes. Due to the different datasets containing metadata such as age of surgery and whether the patient had a history of smoking. We could look at the exact details for the surgery as our intervention and use the dead or alive category as the responder/non-responder property based on the type of surgery and taking into consideration the severity of the condition.
PMID 38935373
Liver Cancer
PMID 34975338
Ovarian Cancer
PMID 35671108
Esophageal Cancer
PMID 37313409
Single Cell RNA Sequencing Data: This paper selects transcriptomic data from the GSE78220 cohort, which included melanoma patients who received anti-PD-1 checkpoint inhibition therapy before treatment.