GW-FEAST Data De-identification: Difference between revisions
Lorikrammer (talk | contribs) mNo edit summary |
mNo edit summary |
||
| (4 intermediate revisions by one other user not shown) | |||
| Line 2: | Line 2: | ||
== Introduction == | == Introduction == | ||
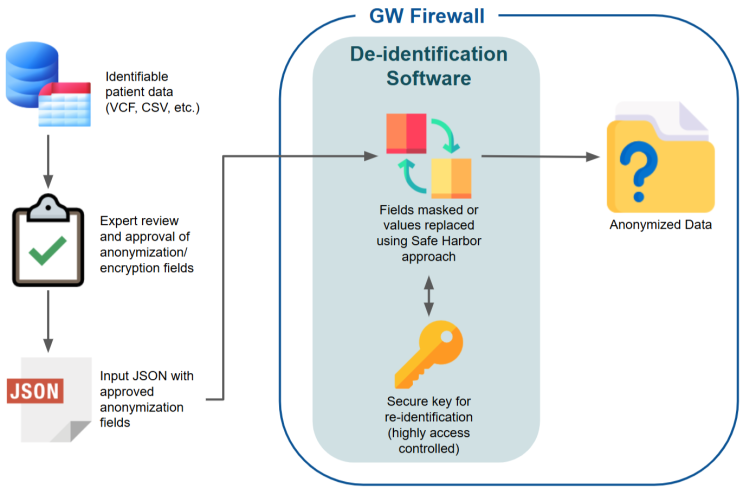
All GW-FEAST datasets housed within the secure data environment are de-identified. This process varies slightly between data sources | All GW-FEAST datasets housed within the secure data environment are de-identified. This process varies slightly between data sources; each has its own versioned de-identification tool and protocol. Our implementation follows the ''Safe Harbor'' recommended deidentification protocol with modifications based on the ''Expert Determination'' method. This de-identification workflow is illustrated below. | ||
=== De-identification Workflow === | === De-identification Workflow === | ||
[[File:Nbcc deidn tool v1.0.png|frameless|743x743px]] | [[File:Nbcc deidn tool v1.0.png|frameless|743x743px]] | ||
=== | === De-identification Data Templates === | ||
Single-patient data templates for each data source are available for download on the [[GW-FEAST De-identified Data Templates]] page. | Single-patient data templates for each data source are available for download on the [[GW-FEAST De-identified Data Templates]] page. | ||
== GWDC De-identification Tool == | == GWDC De-identification Tool == | ||
The GW Data Commons (GWDC) de-identification tool is used to de-identify all GW | The GW Data Commons (GWDC) de-identification tool is used to de-identify all [[GW Data Commons (GWDC) Data|GWDC]] datasets prior to harmonization. This protocol was authored by Dr. Robel Kahsay of the Mazumder Research Group. More information is available on the [[GWDC De-identification Tool]] page. | ||
== NBCC De-identification Tool == | == NBCC De-identification Tool == | ||
The National Breast Cancer Coalition de-identification tool is used to de-identify all NBCC datasets prior to harmonization. This protocol was authored by Dr. Robel Kahsay of the Mazumder Research Group and v1.0 of the tool has been approved by a representative at NBCC. Each iteration of the tool will have written approval by NBCC before use. More information is available on the NBCC | The National Breast Cancer Coalition de-identification tool is used to de-identify all NBCC datasets prior to harmonization. This protocol was authored by Dr. Robel Kahsay of the Mazumder Research Group and v1.0 of the tool has been approved by a representative at NBCC. Each iteration of the tool will have written approval by NBCC before use. More information is available on the [[National Breast Cancer Coalition (NBCC) Data|NBCC Data]] page. | ||
Latest revision as of 11:56, 21 March 2025
← Go Back to the GW-FEAST Home Page.
Introduction
All GW-FEAST datasets housed within the secure data environment are de-identified. This process varies slightly between data sources; each has its own versioned de-identification tool and protocol. Our implementation follows the Safe Harbor recommended deidentification protocol with modifications based on the Expert Determination method. This de-identification workflow is illustrated below.
De-identification Workflow
De-identification Data Templates
Single-patient data templates for each data source are available for download on the GW-FEAST De-identified Data Templates page.
GWDC De-identification Tool
The GW Data Commons (GWDC) de-identification tool is used to de-identify all GWDC datasets prior to harmonization. This protocol was authored by Dr. Robel Kahsay of the Mazumder Research Group. More information is available on the GWDC De-identification Tool page.
NBCC De-identification Tool
The National Breast Cancer Coalition de-identification tool is used to de-identify all NBCC datasets prior to harmonization. This protocol was authored by Dr. Robel Kahsay of the Mazumder Research Group and v1.0 of the tool has been approved by a representative at NBCC. Each iteration of the tool will have written approval by NBCC before use. More information is available on the NBCC Data page.